Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
Computational Analysis and Molecular Docking Approach for Liver Cancer
Authors: Uma Kumari, Kartik Tripathi
DOI Link: https://doi.org/10.22214/ijraset.2023.54927
Certificate: View Certificate
Abstract
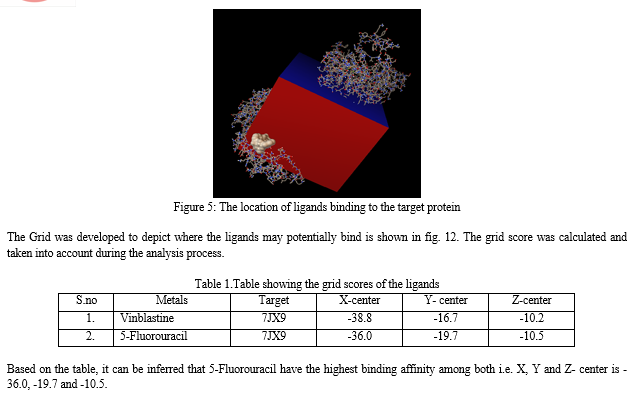
Liver cancer is a serious worldwide medical concern, with death rates that are alarmingly higher than those of other types of cancer. It kills roughly 700,000 people annually, necessitating immediate attention and effective strategies for prevention, diagnosis, and treatment. Hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA) are the most prevalent aggressive liver tumors, with uncommon variants such as fibrolamellar carcinoma and hepatoblastoma contributing to the disease burden. Computational analysis and molecular docking techniques are valuable tools in studying liver cancer. In this study, research was conducted via molecular docking analysis using the 3D structure of human ornithine aminotransferase with a bound intermediate molecule (PDB ID: 7JX9). This approach allows for the investigation of potential interactions between the target protein and ligands. Two ligands, Vinblastine and 5-Fluorouracil, were selected for molecular docking against the target protein. Vinblastine, derived from the plant Vincarosea Linnaeus, is an anticancer alkaloid that hinders microtubule development by interacting with tubulin. 5-Fluorouracil, a synthetic compound resembling the nucleobase uracil, functions as an antimetabolite and inhibits tumor growth. The docking analysis revealed that both ligands were able to bind to the target protein, and specific amino acid residues within the A-domain were identified as involved in the binding of Vinblastine. The docking results were visualized and analyzed using PyMol software. The analysis provided insights into the interactions between the target protein and ligands, shedding light on potential binding sites and affinity. The grid scoring was used in AutodockVina\'s docking procedure to determine how well the protein receptor might bind to the ligands. The calculated grid scores from the analysis showed that Vinblastine (with x-center = -38.8, y-center = -16.7 and z-center = -10.20) and 5-Fluorouracil (with x-center = -36.0, y-center = -19.7, z-center = -10.5).It was seen that the ligand with the lowest grid score is 5-Fluorouracil, with an x-center = -36.0, y-center = -19.7, z-center = -10.5.A ligand\'s binding energy is directly proportional to its grid score, meaning that a lower score indicates a higher binding energy. If the ligand has a lower grid score, it is predicted to have a stronger affinity for the protein.
Introduction
I. INTRODUCTION
Liver cancer is accounting for a disproportionate share of the disease's overall toll as it is one of the major causes of cancer related mortality. Notably, the prevalence and fatality rates of liver cancer are increasing, posing a mounting challenge. About 700,000 people lose their lives to liver cancer every year [1,2]. This cancer includes a wide variety of malignancies, some of which are more common in specific race, environmental conditions and geographic area than others. Hepatocellular cancer and intrahepatic cholangiocarcinoma are types of this cancer. The great variety of liver malignancies can be broken down into several of these groups. In addition, adolescents are disproportionately affected by two rare types of malignant liver tumors. These are fibrolamellar carcinoma and hepatoblastoma. These particular types of liver cancer present unique characteristics and are typically observed in a distinct age group. With high death rate, HCC is considered as an aggressive type of tumor. It demonstrates rapid growth in both its incidence and mortality rates. Moreover, HCC exhibits a higher prevalence among males than females. Liver cirrhosis is a disease which can be caused by variety of causes, and is usually the precursor to HCC. This diverse disease manifests in individuals affected by liver cirrhosis, which can arise from a multitude of underlying factors [3]. Underlying factors contributing to liver cirrhosis, and subsequently hepatocellular carcinoma (HCC), encompass a range of causes. These include viral infections such as HBV and HCV, excessive alcohol consumption, as well as metabolic syndrome. These various etiological factors significantly contribute to the development and progression of liver-related diseases.ICC data related to incidents and death is also increasing globally, making it the second most frequent form of liver cancer. The ethnic groups especially Hispanics and Asians possessed the greatest rates of this tumor type [4]. Both types of tumor share a relationship with persistent liver damage, although having different etiology.
Various chronic liver diseases contribute to the ongoing damage and turnover of liver cells, creating an environment prone to errors during the repair process. Constant cell death and replacement drastically alters the liver's microenvironment, creating a setting where cancer can flourish. As a result, an oncogenic microenvironment emerges, facilitating the progression of both primary liver tumors [5]. Several different approaches can be taken in the case of early tumor detection. Surgical resection, regional ablation procedures, and organ transplantation are all viable options. However, as cancer develops into later stages, fewer therapeutic options are available. Although overall outcomes are mediocre, alternative therapy techniques have shown some positive increases in patient survival [6,7,8]. In depth analysis of liver cancer has unveiled that mature liver cells are main causes of this cancer. Several processes may contribute to the emergence of HCC. In one scenario, hepatocytes are transformed into HCC as a result of the progressive accumulation of specific genetic mutations. De-differentiation of hepatocytes into the precursor cells is another route that can lead to HCC through genetic changes [9].
HCC is characterized by significant mutations in genes that have been shown to drive the disease forward. Recurrent abnormalities in genetics in HCC include those in genes related to cell cycleand i.e. cyclin D1 and cyclin-dependent kinase inhibitor 2A, as well as in genes that are involved in progession of cancer including TP53, MYC, WNT, and beta-catenin [10,11]. Particular genes that show recurrent somatic alterations are suspected of playing a role in carcinogenesis. Hepatocellular carcinomas typically have an average number of alterations per genomes that is roughly the same as that of other cancers. In addition, HBV RT doesn't have a proofreading function, which likely contributes to the emergence of mutations. However, HCV infection causes dsDNA breaks, which contributes to a higher mutation rate. Mutations in some genes are more common in HCV-infected cells [12,13].
Telomerase is an assembly of proteins made up of the reverse transcriptase (RT) and RNA portion of telomerase, and it plays an important role in preserving the stability and longevity of telomeres. Damage to DNA response is triggered after telomeres are significantly shorter [14]. Either apoptosis or senescence of cells can result from this reaction. This process is harmful to the functioning of the liver because it contributes to inflammation and, eventually, cirrhosis by preventing the liver from renewing its default structure [15]. A total of 35 prognostic and immune-related genes (PRGs) for HCC were discovered in 2022. The survival rate was noticeably better in the low-risk subgroup than in the group with the greatest risk. Immunological-related high-risk populations were shown to have lower immune status in investigations of the Genetic Ontology and Kyoto's Encyclopedia for Gene and Genome. PRGs are very relevant to cancer immunity and may be used as a prognostic indicator for folks with HCC [16]. In a study conducted in 2021 with a focus of this investigation was to explore the intricate relationship between long non-coding RNAs (lncRNAs) and the genes associated with ferroptosis, a form of regulated cell death, in HCC. The study encompassed a comprehensive analysis of HCC samples and samples of normal liver tissue. By exploring the interplay between lncRNAs and ferroptosis-related genes, the researchers revealed novel insights into the potential diagnostic and prognostic capabilities of these non-coding RNA molecules [17]. In bioinformatics analysis conducted in 2021, researchers successfully identified six crucial genes that hold significant promise for the early detection and treatment of liver cancer. The study involved the evaluation of a total of 1564 genes, from which they observed that 1400 genes exhibited increased activity, while 164 genes displayed decreased activity. Through careful verification and analysis, six genes emerged as particularly noteworthy [18]. Colorectal cancer metastatic to the liver candidate fundamental genes and important genes were found using bioinformatics research in 2021. The comparison of CRC patients with liver metastases using four datasets revealed 35 DEGs (set 1) and 142 DEGs (set 2). Following the development of the PPI network, researchers identified ITIH2 as a candidate main gene for liver metastasis in colorectal cancer and COL1A2 as a crucial gene in the development of colorectal carcinoma [19]. In 2020, m6A methylation-related genes from The Cancer Genome Atlas (TCGA) were used to predict outcomes in HCC patients. Bioinformatics were used to decipher the predictive significance of transcriptomic and clinical data for 15 genes. Overexpression of 11 genes in HCC was associated with decreased survival. Patients with high risk were also found to have altered expression of ZC3H13, while METTL3 and KIAA1429 etc. were all down-regulated. The study emphasizes the variations in m6A methylation-related gene expression patterns among HCC patients [20,21,22,23].
II. MATERIALS AND METHODS
The National Institutes of Health (NIH) in the USA administers the National Center for Biotechnology Information (NCBI) as one of its organizations. The National Center for Biotechnology Information (NCBI) is an internet platform for the dissemination of medical data and bioinformatics software that uses computational methods to answer fundamental biological questions [24,25]. X-ray crystallography, NMR spectroscopy, and electron microscopy, all of which identify three-dimensional structures, are all made available here. Researchers from several fields, including biochemistry, biophysics, structural biology, drug development, and molecular modeling, make extensive use of the PDB to further improve comprehension of the structure and function of molecules in biology [26,27].
RasMol is a free and open-source application for showing and analyzing biological molecules, especially proteins, nucleic acids, and tiny molecules. It also provides a collaborative setting for analyzing and altering 3D molecular structures. RasMol can read molecular coordinates from numerous file formats, including PDB, Mol2, CHARMm, and mmCIF, and it supports a variety of methods for representation, including wireframe, space-filling, ribbon, and illustration representations [28]. Warren Lyford DeLano created and initially disseminated PyMOL, a freely accessible software system for viewing macromolecules. The software utilizes the Python programming language and supports numerous molecular visualization formats, including ribbon, caricature, spots, surface, spheres, stick, and line [29,30,31]. AutoDockVina is widely utilized and swift open-source molecular docking software. Along with AutoDock4 (AD4), AutoDock-GPU, AutoDock-FR, and AutoDock-CrankPep, it is one of the docking tools included in the AutoDock Suite. Vina is regarded as one of the most widely used applications due to its simplicity and efficiency in comparison to other docking programs, both within and beyond the AutoDock Suite. In addition, the program is open-source. It is a set of tools used for automating docking, which attempts to anticipate how small molecules, such as substrates or potential drugs, will bind to a 3D-structured receptor [30].
III. RESULT AND DISCUSSION
For conducting computational analysis and molecular docking, the required target information was obtained from the National Center for Biotechnology Information (NCBI) database, including the PDB ID (7JX9). Subsequently, the 3D structure of the target was downloaded in PDB format from the Protein Data Bank (PDB) database. In this project, the target is identified as 7JX9, which is a three-dimensional arrangement of human ornithine aminotransferase with a bound intermediate molecule while undergoing inactivation caused by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid.
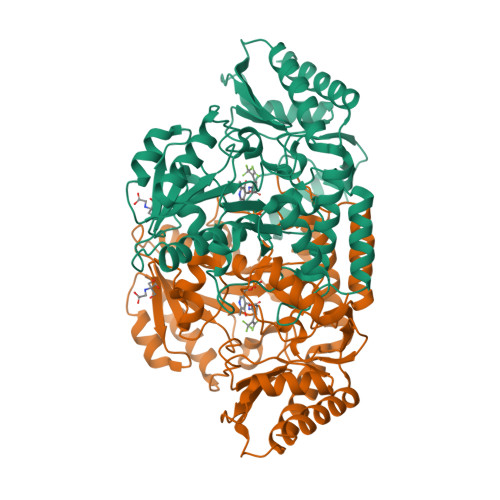
Figure 1: 3D structure of 7JX9 from PDB
In the subsequent step, two ligands, Vinblastine and 5- Fluorouracil, were chosen for molecular docking against the target protein. Based on the literature review, it can be inferred that 5-fluorouracil is a synthetic compound that resembles the nucleo-base uracil, with the substitution of a hydrogen atom at position 5 with fluorine. This compound functions as an antimetabolite and is classified as an agent that can be used in the treatment of cancer by slowing down the growth of tumor. Similarly, the organic compound vinblastine is derived from a plant called Vincarosea Linnaeus. It is referred to as an anticancer alkaloid that hinders the development of micro-tubule by interacting with tubulin. This interference interrupts the formation of the spindle at mitosis and forces tumor cells to move into an M-phase arrest in their cell cycle.
Next, Vinblastin and 5- Fluorouracil were required as ligands. To obtain these structures, PubChem database was accessed and downloaded the structures of these metals. These structures will be utilized in the later stages of the project. In order to study the structure of the target molecule, we utilized the RasMol software. This software allows us to read molecular coordinates from a file and interactively display the molecule in various representations. Specifically, we used RasMol to analyze the structure of target 7JX9 and gain a deeper understanding of its molecular composition. After launching RasMol, the target structure was loaded into the software. This was achieved by importing the relevant file from our computer into RasMol.By selecting Command Line window, a new interface was displayed that allowed us to interact with the loaded structure using a series of commands. Once the structure was loaded, investigation of target properties and characteristics by issuing commands and using the software's tools and functions was done.
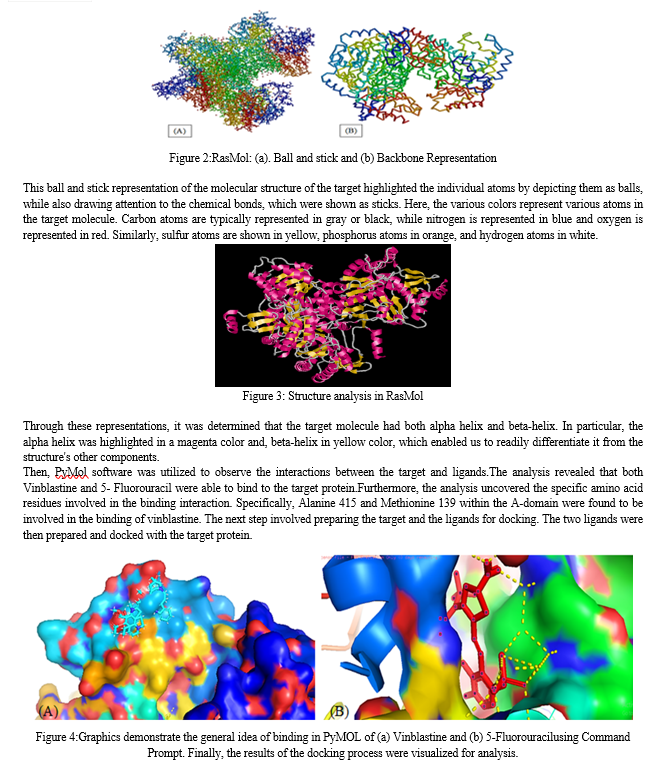
Conclusion
In conclusion, liver cancer represents a significant global health challenge with high mortality rates. The molecular analysis and docking studies conducted in this research contribute to our understanding of liver cancer at a molecular level and offer potential insights for the development of targeted therapies. The findings underscore the importance of early detection, genetic analysis, and computational approaches in advancing liver cancer research and improving patient outcomes. Further investigations and clinical trials are warranted to validate the efficacy of identified ligands and explore their therapeutic potential in the treatment of liver cancer.
References
[1] Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017 May 29;16:1. [2] Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016 Apr 14;2:16018. [3] Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017 Mar;152(4):745-761. [4] Massarweh NN, El-Serag HB. Epidemiology of Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Control. 2017 Jul-Sep;24(3):1073274817729245. [5] Chidambaranathan-Reghupaty S, Fisher PB, Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. 2021;149:1-61. [6] Tabori NE, Sivananthan G. Treatment Options for Early-Stage Hepatocellular Carcinoma. SeminInterventRadiol. 2020 Dec;37(5):448-455. [7] Wells SA, Hinshaw JL, Lubner MG, Ziemlewicz TJ, Brace CL, Lee FT Jr. Liver Ablation: Best Practice. RadiolClin North Am. 2015 Sep;53(5):933-71. [8] Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S, Lencioni R, Koike K, Zucman-Rossi J, Finn RS. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021 Jan 21;7(1):6. [9] Kumar M, Zhao X, Wang XW. Molecular carcinogenesis of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: one step closer to personalized medicine? Cell Biosci. 2011 Jan 24;1(1):5. [10] Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol. 2016 Nov 7;22(41):9069-9095. [11] Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. 2017 Aug 30;16(1):149. [12] Goerlitz DS, Blancato J, Ramesh A, Islam M, Graham GT, Revina V, Kallakury B, Zeck J, Kirillova E, Loffredo CA. Somatic mutation signatures in primary liver tumors of workers exposed to ionizing radiation. Sci Rep. 2019 Dec 3;9(1):18199. [13] Ng SWK, Rouhani FJ, Brunner SF, Brzozowska N, Aitken SJ, Yang M, Abascal F, Moore L, Nikitopoulou E, Chappell L, Leongamornlert D, Ivovic A, Robinson P, Butler T, Sanders MA, Williams N, Coorens THH, Teague J, Raine K, Butler AP, Hooks Y, Wilson B, Birtchnell N, Naylor H, Davies SE, Stratton MR, Martincorena I, Rahbari R, Frezza C, Hoare M, Campbell PJ. Convergent somatic mutations in metabolism genes in chronic liver disease. Nature. 2021 Oct;598(7881):473-478. [14] O\'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010 Mar;11(3):171-81. [15] Calado RT, Brudno J, Mehta P, et al. Constitutional telomerase mutations are genetic risk factors for cirrhosis. Hepatology. 2011;53(5):1600–1607. [16] Jiang Y, Sun A, Zhao Y, Ying W, et al., Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019 Mar;567(7747):257-261. [17] Xu Z, Peng B, Liang Q, Chen X, Cai Y, Zeng S, Gao K, Wang X, Yi Q, Gong Z, Yan Y. Construction of a Ferroptosis-Related Nine-lncRNA Signature for Predicting Prognosis and Immune Response in Hepatocellular Carcinoma. Front Immunol. 2021 Sep 17;12:719175. [18] Wang YC, Tian ZB, Tang XQ. Bioinformatics screening of biomarkers related to liver cancer. BMC Bioinformatics. 2021 Oct 25;22(Suppl 3):521. [19] Niu L, Gao C, Li Y. Identification of potential core genes in colorectal carcinoma and key genes in colorectal cancer liver metastasis using bioinformatics analysis. Sci Rep. 2021 Dec 14;11(1):23938. [20] Liu J, Sun G, Pan S, Qin M, Ouyang R, Li Z, Huang J. The Cancer Genome Atlas (TCGA) based m6A methylation-related genes predict prognosis in hepatocellular carcinoma. Bioengineered. 2020 Dec;11(1):759-768. [21] He D, Zhang X, Tu J. Diagnostic significance and carcinogenic mechanism of pan-cancer gene POU5F1 in liver hepatocellular carcinoma. Cancer Med. 2020 Dec;9(23):8782-8800. [22] Song X, Du R, Gui H, Zhou M, Zhong W, Mao C, Ma J. Identification of potential hub genes related to the progression and prognosis of hepatocellular carcinoma through integrated bioinformatics analysis. Oncol Rep. 2020 Jan;43(1):133-146. [23] Deng M, Sun S, Zhao R, Guan R, Zhang Z, Li S, Wei W, Guo R. The pyroptosis-related gene signature predicts prognosis and indicates immune activity in hepatocellular carcinoma. Mol Med. 2022 Feb 5;28(1):16. [24] Virk N, Kumari U. Genome Sequence Analysis of Lungs Cancer Protein WDR74 (WD Repeat-Containing Protein). IJRASET. 2022;10(V):4533-4537. [25] Sayers EW, Agarwala R, Bolton EE, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2019;47(D1):D23-D28. [26] NK Virk et al., Identification of New Potential Drugs for Lung Adenocarcinoma Causing Protein RMB10 Using Computer-Aided Drug Design Approach. International Journal of Bio-Technology and Research (IJBTR). 2022;12(2):1-8. [27] Berman HM, Westbrook J, Feng Z, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28(1):235-242. [28] Pikora M, Gieldon A. RASMOL AB - new functionalities in the program for structure analysis. ActaBiochim Pol. 2015;62(3):629-31. [29] Yuan S, Chan HCS & Hu Z. Using PyMOL as a platform for computational drug design. Wiley Interdisciplinary Reviews: Computational Molecular Science. 2017;7(2):1-10. [30] Dr Uma kumari, Devanshi Gupta, In silico RNA aptamer drug design and modelling,2022/4, Journal-JETIR,Volume-9, Issue-4, Pages 718-725 [31] Uma kumari, NavjotKaurVirk, Identification of new potential drug for lung adenocarcinoma causing protein RMB10 using computer aided drug design approach, Publication date- 2022/6/11, Journal-IJBTR, Volume-12, Issue-2, Pages 1-8 [32] Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017;9(2):91-102.
Copyright
Copyright © 2023 Uma Kumari, Kartik Tripathi. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET54927
Publish Date : 2023-07-23
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

